CRISPR-Cas9 gene editing is a powerful technology that allows researchers to introduce precise changes to DNA sequences within cells. Also, as these changes are permanent, they are inherited by the cells’ progeny.
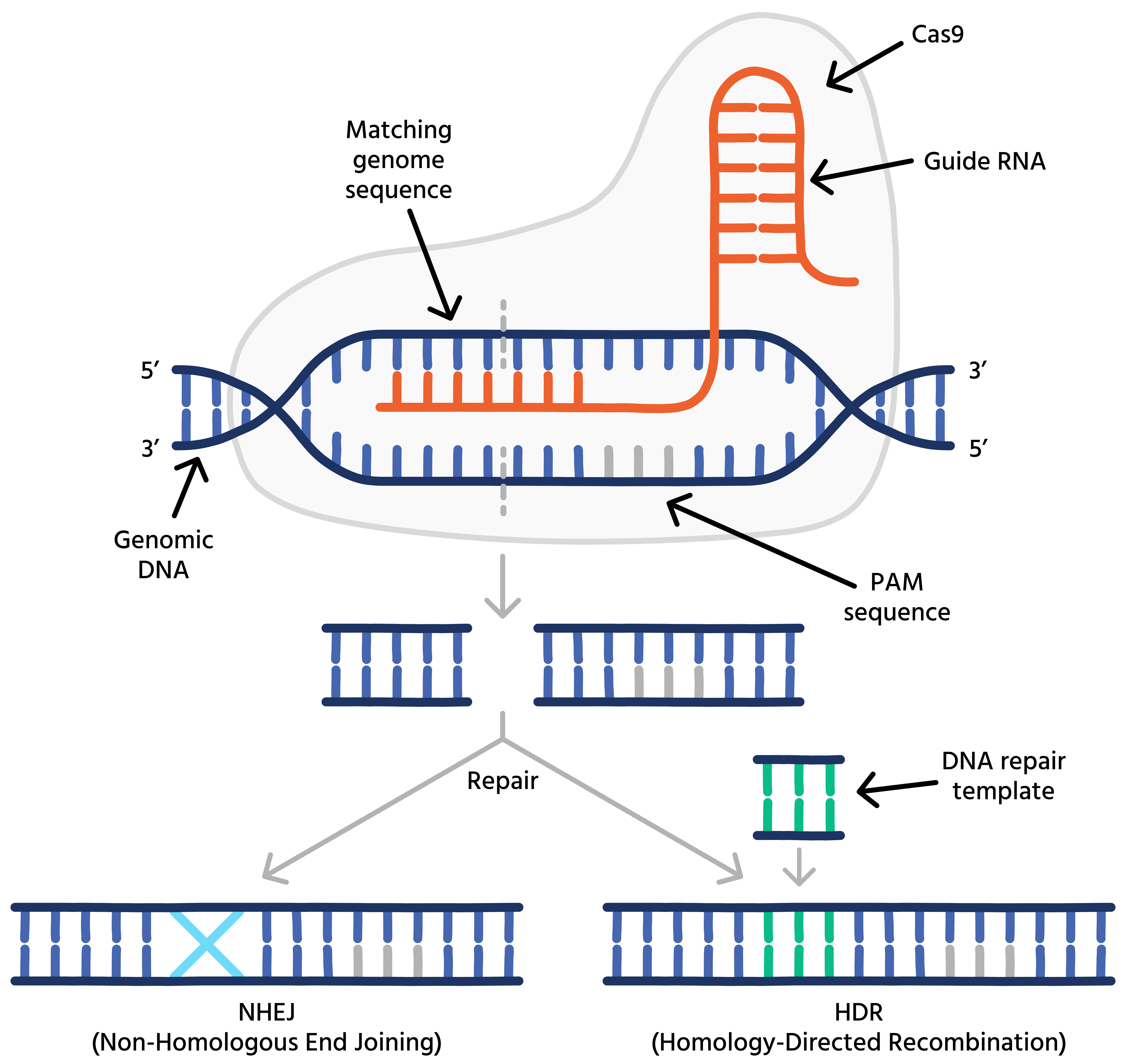
Although not the first gene-editing technology, CRISPR-Cas9 is much simpler compared to other approaches like Zinc-fingers and TALENs. It introduces double-stranded breaks (DSBs) using a wild-type Cas9 protein and a guide RNA, forming a ribonucleoprotein (RNP) complex. This complex recognizes its designated genomic target through sequence homology between the guide RNA and genomic DNA. Therefore, simply switching guide RNAs allows scientists to target Cas9 to different genomic locations.
For a DSB to occur, a protospacer-adjacent motif (PAM) must be present near the targeted sequence. For Cas9, the PAM is 5′-NGG’-3′. Once a DSB is introduced, cells repair the damage through various mechanisms. These mechanisms can be exploited to create functional gene knockouts, precise edits of individual bases, or to introduce tags into the genome.
The first naturally occurring clustered interspaced short palindromic repeats (CRISPR) sequence was identified in the bacterium E.coli and published in 1987 by Ishino et al.. However, researchers coined the term ‘CRISPR’ much later and the relevance of these curious sequence arrangements remained unknown for many more years.
Subsequently, scientists recognized the significance of CRISPR and Crispr-associated genes (Cas) in bacterial immune defence (Mojica et al., 2005, Barrangou et al., 2007).
In 2012, Jinek et al. demonstrated that CRISPR-Cas9 could be programmed to target specific DNA sequences, thus paving the way for a new era of precise gene-editing.
Jennifer A. Doudna, and Emmanuelle Charpentier were awarded the Nobel price in Chemistry in 2020 ‘”for the development of a method for genome editing”.

Successfully establishing gene-edited cells entails several steps, including creating a pool of gene-edited cells, single-cell cloning to separate edited from non-edited cells, screening of clones for the desired edit, as well as upscaling and banking of selected clones for downstream applications. According to our recent survey about CRISPR-Cas9, single-cell cloning poses the biggest challenge of the workflow. iotaSciences’ Cloning Platform greatly simplifies the single-cell cloning process by automating all tedious liquid handling steps, while reliably assuring monoclonality utilizing miniature cell-culture chambers.
After designing guide RNAs and repair templates, deliver the editing reagents to target cells using plasmid-based approaches or RNP complexes (Cas9 protein and guide RNAs). The choice of transfection methods, such as electroporation or lipid-mediated delivery, depend on cell compatibility and the cargo to deliver. Viral-mediated delivery is also an option. Following a brief recovery period after transfection, assess editing efficiency in the pool using targeted PCR followed by Sanger sequencing and Inference of CRISPR Edits (ICE) analyses.
After establishing a CRISPR-Cas9 edited cell pool, it's crucial to isolate single cells and grow them into clonal cultures. The initial pool contains both edited and non-edited cells, making this separation step essential. Traditional manual single-cell cloning methods are tedious and unreliable for ensuring monoclonality.
Our automated Cloning Platform solves these problems by using miniature culture chambers for easy and reliable monoclonality verification. It also automates all pipetting tasks, streamlining the process and ensuring consistency.
Once clonal lines are established, assess each for the desired edit. The number of clones to test depends on the initial cell pool's editing efficiency (see step 1) and the number of edited clones needed for downstream applications. Replica-plating colonies across multiple 96-well plates is very useful at this stage: use one plate for analysis and keep the other one as a backup. Identify edited clones using targeted PCR on genomic DNA from the clones, followed by Sanger sequencing. The genomic DNA can further be used to check for potential off-target edits in selected clones.
The selected edited clones need further upscaling. For this, transfer the clones to larger vessels and continue culturing them until they can be frozen and stored in several cryovials per clone. These initial vials form the master cell bank (MCB) which provides a supply of genetically and phenotypically homogenous cells with a lower passage number. A working cell bank (WBC) can optionally be created to further upscale. The newly generated cell models can now be used for downstream applications, such as disease modelling, drug screening, organoid culture and many others.
Want to find out more about our single-cell platforms? Contact one of our knowledgeable experts today, visit our resources page, or request a demo to see the platforms in action.