CRISPR-Cas9 gene editing - survey report
CRISPR-Cas9 gene editing has been revolutionizing the scientific landscape ever since its discovery and positioning as a gene editing tool. It allows scientists to change (“edit”) genomic DNA sequences within cells precisely. The ease-of-use compared to other gene editing technologies, such as Zinc-fingers and TALENs, has significantly accelerated diverse life science disciplines.
This 2024 survey report offers insights into applications, methods and associated challenges, by compiling responses from diverse researchers and labs. We summarize key patterns, highlighting diversity and emerging practices.
A massive thank you to all our 150+ survey participants for sharing their knowledge and experiences with the community!
- Participants - overview
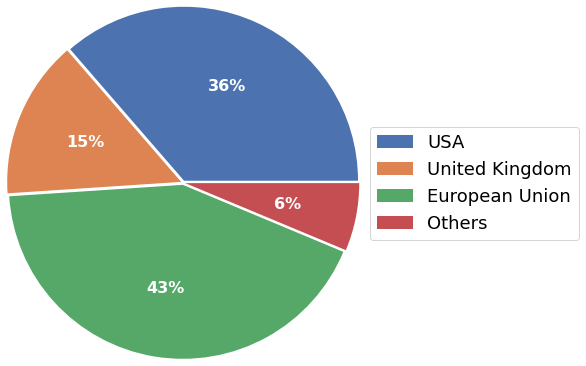
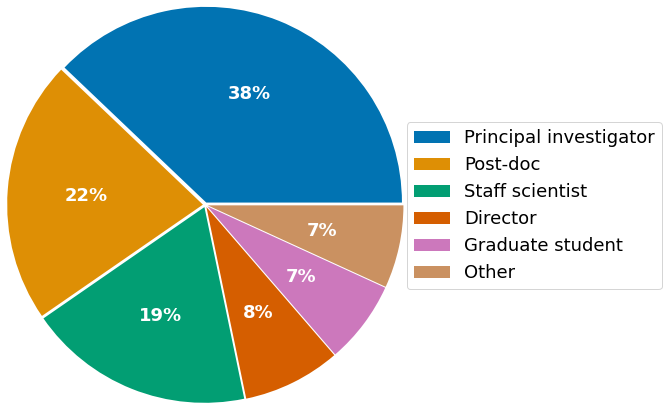
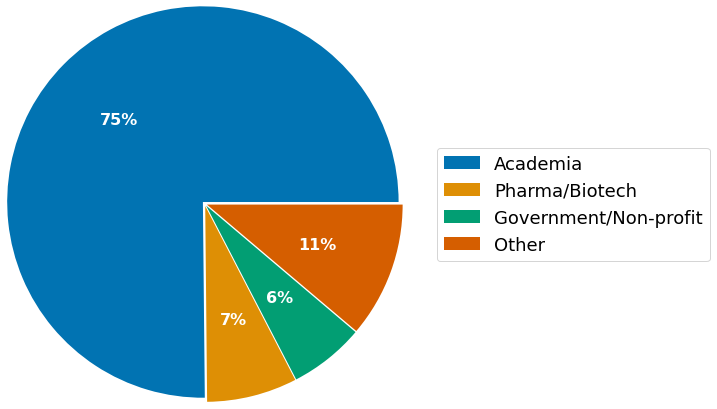
The survey presents answers from highly experienced scientists in senior positions, with the largest fraction represented by PIs (38%), followed by Post-docs (22%), and staff scientists (22%). Mostly, participants affiliate with academia (75%), with the majority of respondents currently working in the European Union (43%), followed by the US (36%), and the United Kingdom (15%).
- Work location
- Current role
- Affiliation
- Technologies and applications
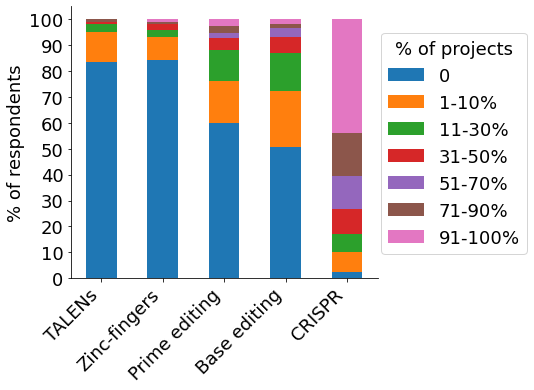
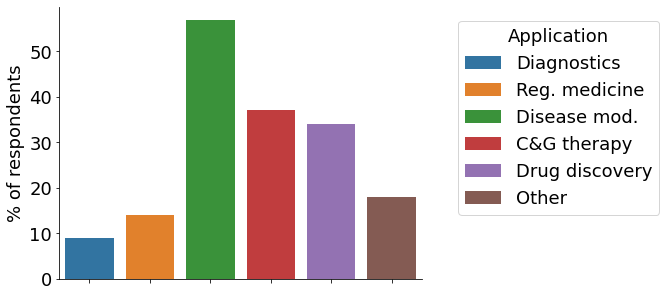
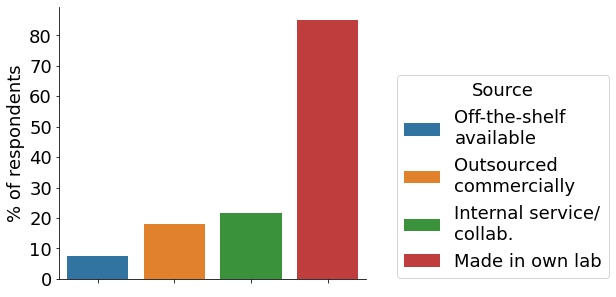
Our participants are using a variety of gene-editing technologies for their research, with CRISPR being the most utilised approach. Almost half of participants (~45%) have used CRISPR for 91-100% of their gene-editing projects (“CRISPR”, pink bar), with <5% indicating to not have used it for any project (blue bar). More than half apply the technologies for disease modelling, followed by cell & gene therapy research and drug discovery. A sizeable fraction of scientists indicated “other” applications not represented in the survey. At large, cell models are generated in the own lab, followed by internal services & collaborators, as well as commercially outsourced.
- Applied technology
- Research Applications
- Source of engineered cells
- CRISPR experiences and challenges
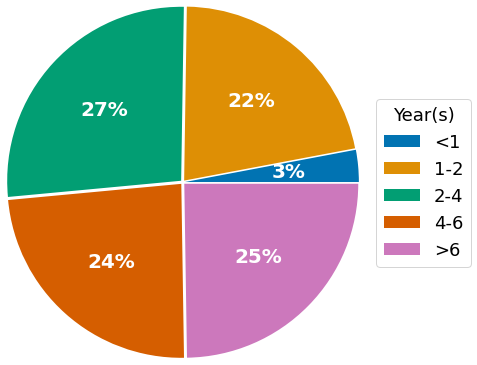
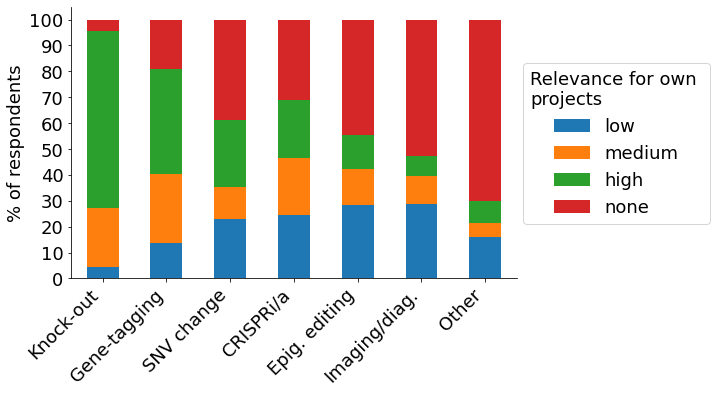
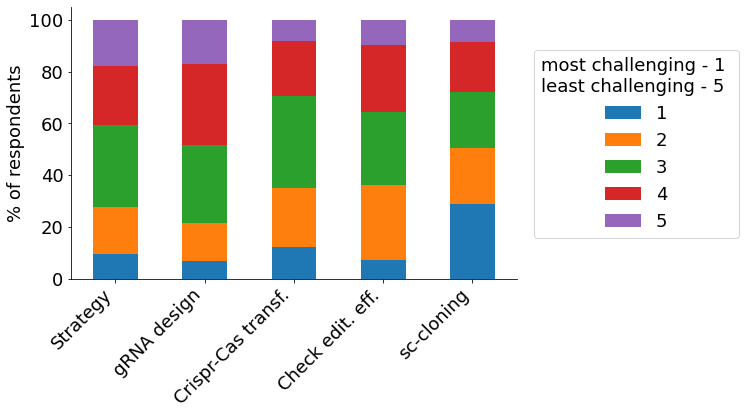
Survey participants are highly experienced with CRISPR, with 25% possessing 6+ years of experience, and ~75% using it for more than two years. The most relevant use case of CRISPR is the creation of knock-outs, with more than 90% of survey respondents indicating it to be of high or medium relevance for their research (“Knock-out”, green & orange bar). Among the various steps in the CRISPR gene-editing workflow, single-cell cloning is considered the biggest challenge, with about half of survey respondents rating it 1 or ‘2 out of 5 (1 – most challenging, 5 – least challenging).
- Years experience
- Use case
- Challenges
- Cell models
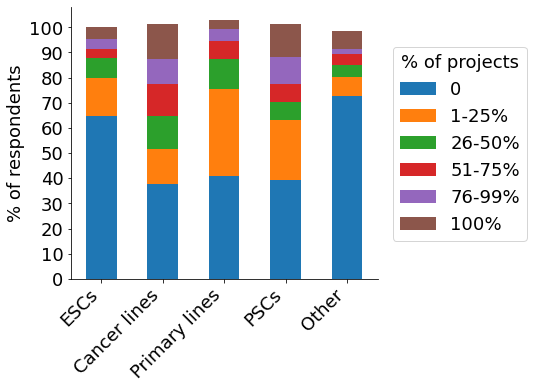
Cancer cell lines and pluripotent stem cells are the top cell types used among our participants, with more than 10% of respondents indicating that all their CRISPR projects utilise them (‘Cancer lines’, ‘PSCs;, brown bar). The majority of participants establish between 1-5 gene-edited cell lines per year (orange & green bar). Roughly a third of scientists indicated engineering 6+ cell lines per year, with some labs creating 20+. Around half of researchers require between 3-4 months to engineer a cell model (orange bar).
- Cell types
- Engineered cell lines per year
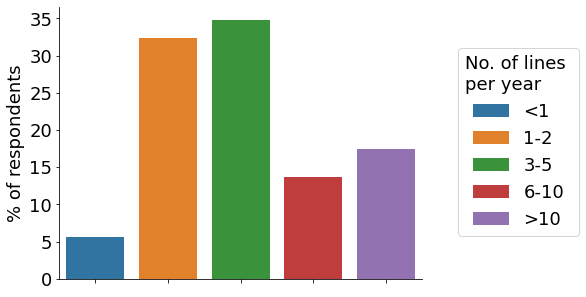
- Time to engineer cell model
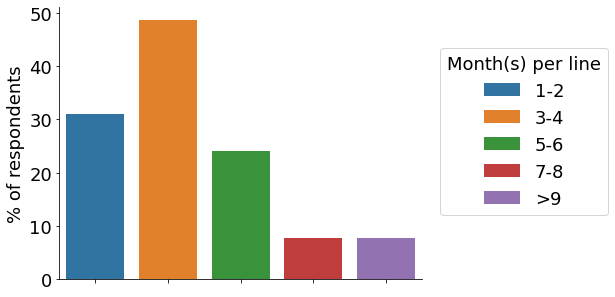
- CRISPR- gRNAs & efficiency
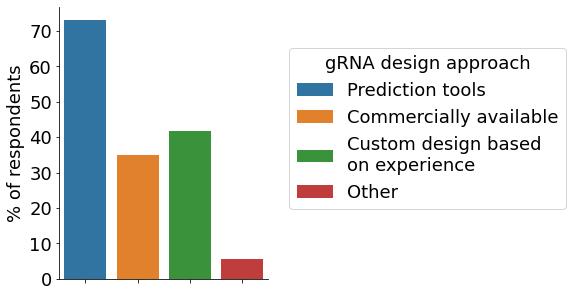
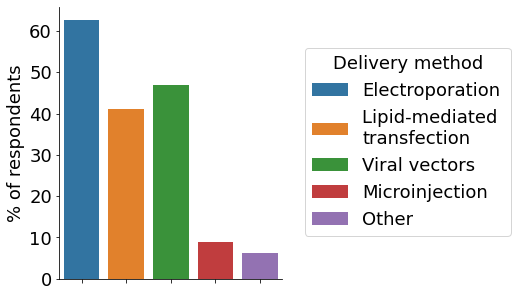
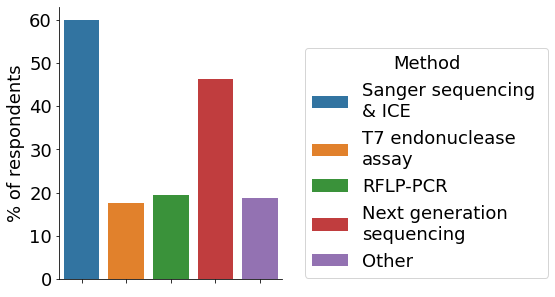
The initial phase of a CRISPR project comprises the design of gRNAs. For this, the majority of survey participants utilise in-silico prediction tools, followed by designing custom gRNAs based on previous experiences (multiple choices allowed). The most popular approach for delivering CRISPR components to cells is electroporation, followed by viral vectors and lipid-mediated transfection. For the initial check on editing efficiency multiple approaches exist. In our survey, Sanger sequencing and Inference of CRISPR Edits (ICE), followed by NGS were the most employed ones.
- gRNA design approach
- Reagent delivery to cells
- Assessing editing efficiency
- CRISPR - off-targets
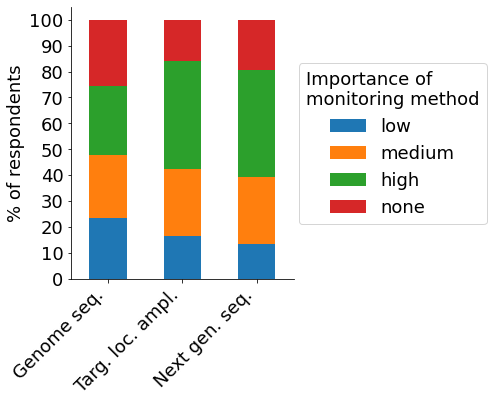
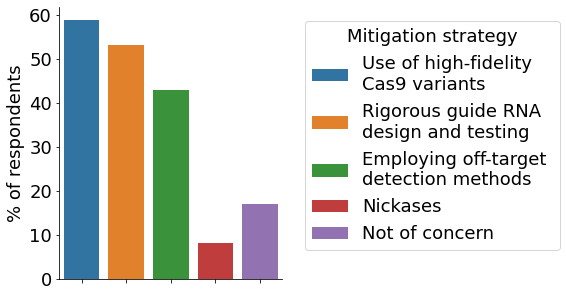
An ongoing concern in the field of CRISPR research are off-target effects that change untargeted DNA sequences. Targeted locus amplification and NGS are considered the most important methods for monitoring off-targets among our participants. The most popular strategy to mitigate potential off-target effects is the use of high-fidelity Cas9 variants, followed by rigorous gRNA design and the employment of off-target detection methods. Nickases are only employed by a minority of participating scientists, possibly due to the increased complexity involved.
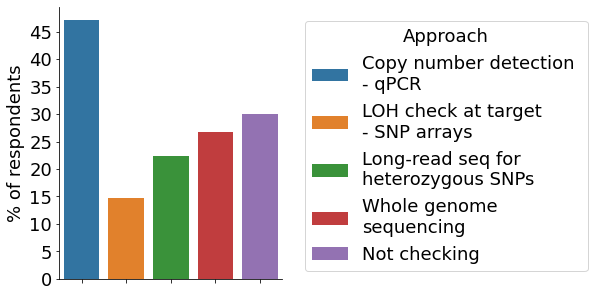
CRISPR may also elucidate unwanted ‘on-target’ effects at the targeted site, including larger deletions. The most common method to assess on-target effects were copy number detection via qPCR, while ~1/3 of respondents indicated to not look for unintended on-target effects.
- Off-target monitoring
- Off-target mitigation
- On-target detection